Project Information and Reproducibility Guide
View the Project on GitHub LabNeuroCogDevel/corticalmyelin_maturation
Heterochronous laminar maturation in the human prefrontal cortex
Project Lead
Valerie J. Sydnor
Project Mentor
Beatriz Luna
Collaborators
Daniel Petrie, Shane D. McKeon, Alyssa Famalette, Will Foran, Finnegan J. Calabro
Project Start Date
September 2023
Current Project Status
Manuscript in submission
Datasets
Luna-7T
Github Repository
https://github.com/LabNeuroCogDevel/corticalmyelin_maturation
Project Directories
The project directory on rhea is: /Volumes/Hera/Projects/corticalmyelin_development
The directory structure on rhea is as follows:
BIDS: BIDS-compliant MP2RAGE data
BIDS/derivatives: Processed image derivatives including unicort-corrected UNI and T1 maps, volumetric R1 maps, and surface measures
Dicoms: Organized dicoms for BIDSifying
EEG: EEG aperiodic activity spreadsheets
Figures: Manuscript figure panels
Maps: Brain maps galore! HCP-MMP atlas, S-A axis, Neurosynth z-score maps, AHBA dense gene expression, cytoarchitectural variation, MWF imaging, and templateflow templates
code: local clone of corticalmyelin_maturation repo
gam_outputs: Gam-derived statistics (developmental_effects, sex_effects, sensitivity_analyses, eeg_associations, cognitive_associations)
sample_info: sample demographics, behavioral data, imaging QC spreadsheets, and final participant lists
software/: Directory with project software installs and containers
The project directory on the PSC is: /ocean/projects/soc230004p/shared/datasets/7TBrainMech
Conference Presentations
- Sydnor, V.J., Petrie, D., McKeon, S.D., Famalette, A., Foran, W., Calabro, F.J., Luna, B. Gradients of myelin growth across layers of human prefrontal cortex: Balancing adolescent circuit plasticity and stability. Poster presented at the Frontal Cortex Gordon Research Conference, Holderness, NH, Aug 2024 best poster award
- Timescales of Myelin Maturation in the Human Prefrontal Cortical Ribbon. Presented at the Organization for Human Brain Mapping 32nd Annual Meeting as part of the Symposium “Quantitative Imaging of Myelin Plasticity”, Brisbane, Australia.
CODE DOCUMENTATION
Below is a detailed walk through of all of the code run for this project, including code for image processing, statistical analysis, results derivation, and visualization! The entire analytic workflow is described and links to the corresponding code on github are provided. Please feel free to reach out to Valerie Sydnor (sydnorvj@upmc.edu) with any questions.
Image Analysis Pipeline: UNI and R1 Processing
Organization and Curation of BIDS
Structural data dicoms from the Luna-7T Brain Mechanisms project were first organized into {subject}/{session} organization (required for heudiconv) with the script /BIDS/BIDS_heudiconv/organize_dicoms_forBIDS.sh. These dicoms were then converted to niftis according to the Brain Imaging Data Structure via heudiconv (version 0.13.1). Specifically, data were BIDSifyed using the /BIDS/BIDS_heudiconv/7TBrainMech_MP2RAGE_heuristic.py heuristic and the /BIDS/BIDS_heudiconv/BIDSify_7T.sh script.
In order to check the acquisition parameters for the structural MP2RAGE data acquired for this study, Curation of BIDS on Disk (CuBIDS) was run with /BIDS/CuBIDS/cubids_acquisition_group.sh.
Processing of MP2RAGE-Derived UNI Images
MP2RAGE-derived Uniform (UNI) images were denoised off of the scanner and then corrected for residual B1+ transmit field biases using SPM’s UNICORT algorithm. UNICORT was applied in /image_processing/B1+_transmitfield_correction/unicort/unicort_inhomogeneitycorrection_UNI.m using the configuration parameters specified in /image_processing/B1+_transmitfield_correction/unicort/unicort_configparams_UNI.m. UNICORT outputs were organized and the B1-corrected image was put into the BIDS directory for use with BIDS apps via /image_processing/B1+_transmitfield_correction/unicort/organize_unicort_files.sh.
Following UNICORT, corrected UNI images were run through FreeSurfer to generate person-specific cortical surface reconstructions to map R1 data to. Images were processed with FreeSurfer’s longitudinal analysis stream (FS version 7.4.1) using a containerized FreeSurfer BIDS App. FreeSurfer was run on the Pittsburgh Super Computer by submitting subject-specific jobs via slurm/sbatch. Jobs were submitted with the script /image_processing/longitudinal_freesurfer/freesurfer_submitjobs_PSC.sh, which calls /image_processing/longitudinal_freesurfer/longitudinal_freesurfer_call.sh. Cross-sectional, template, and longitudinal steps of recon-all were run for all subjects (and sessions). Job completion was checked with /image_processing/longitudinal_freesurfer/check_freesurferjobs_complete.sh.
Processing of MP2RAGE-Derived R1 Maps
MP2RAGE-derived quantitative T1 maps were corrected for B1+ transmit field biases using UNICORT by running /image_processing/B1+_transmitfield_correction/unicort/unicort_inhomogeneitycorrection_T1map.m, which uses /image_processing/B1+_transmitfield_correction/unicort/unicort_configparams_T1map.m as the config file. UNICORT output files were renamed and organized with /image_processing/B1+_transmitfield_correction/unicort/organize_unicort_files_T1map.sh.
UNICORT-corrected quantitative T1 maps were next converted into volumetric R1 maps (s^-1). R1 maps were calculated in /image_processing/R1_surfacemaps/R1_volumetricmaps.sh by coverting the T1 readout from ms to s and then calculating R1 = 1/T1. Volumetric R1 maps were then projected to participant- and timepoint-specific cortical surfaces on the PSC by running /image_processing/R1_surfacemaps/R1_vol2surf_submitjobs_PSC.sh. This script calls /image_processing/R1_surfacemaps/voltosurf_projection_nativefsaverage.sh for every session in order to project R1 data to the surface at 11 projection fractions: 0 (GM/WM boundary), 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1.0 (pial boundary).
Calculation of Depth-Dependent R1, Cortical Thickness, and Cortical Curvative in HCP-MMP Regions
After running FreeSurfer and projecting depth-dependent R1 data to the cortical surface, anatomical metrics (cortical thickness, cortical curvature) and depth-dependent R1 metrics were quantified in individual cortical regions in the HCP-MMP (Glasser) atlas. This was accomplished by running /surface_metrics/fstabulate/fstabulate_R1anat_submitjobs_PSC.sh, which ultimately uses the shell and python scripts in /surface_metrics/fstabulate.
The workhorse script directly called via fstabulate_R1anat_submitjobs_PSC.sh is /surface_metrics/fstabulate/collect_stats_to_tsv.sh. This script parses user-defined inputs and transforms fsaverage parcellations to subject’s FreeSurfer native surfaces, runs FreeSurfer’s qcache + mris_anatomical_stats + mri_segstats to get regional metrics of interest, and converts metric stats files to tsvs. After fstabulate_R1anat_submitjobs_PSC.sh completed, /surface_metrics/surface_measures/collate_study_anatomicalstats.py was run to combine session-specific tsv metric files into study parquet files with all participants’ anatomical and depth-dependent R1 data. These parquets were used to generate final input dataframes for statistical modeling.
Image Analysis Pipeline: Partial Voluming and Unique Voxel Compartments in the Cortical Ribbon
Determining the Cortex Volume Fraction at Each Intracortical Depth
The cortex volume fraction was calculated in each HCP-MMP region at all 11 intracortical depths by first estimating the cortex tissue fraction in volumetric space (i.e., the percent of each voxel occupied by cortex) using FreeSurfer’s mri_compute_volume_fractions, which was applied via /image_processing/cortexfraction_surfacemaps/cortexfraction_volumetricmaps.sh. Volumetric cortex fraction maps were then projected to participant-specific cortical surfaces at the 11 intracortical depths in /image_processing/cortexfraction_surfacemaps/cortexfraction_vol2surf.sh and the average cortex fraction in each region at each depth was computed with /surface_metrics/fstabulate/fstabulate_cortexpve_submitjobs_PSC.sh followed by /surface_metrics/surface_measures/collate_cortexpve_stats.py. This allowed for the identification of 7 (of 11) intracortical depths where >90% of the R1 signal arose from corical gray matter, and thus for the study of depths with minimal partial voluming.
Identifying Unique Voxel Compartments in the Cortical Ribbon
We determined the number of unique voxels contained within the cortical ribbon at all frontal lobe vertices using R1 maps from all participants. First, all voxels in the frontal lobe of volumetric R1 maps were assigned unique integer values, and these values were projected to participant- and timepoint-specific cortical surfaces at the same 11 projection fractions used in the main R1 analysis. This was accomplished via /image_processing/corticalribbon_voxelcounts/corticalribbon_voxel_mapping.sh. The number of independent voxel values present across the 7 analyzed depths was computed in /image_processing/corticalribbon_voxelcounts/corticalribbon_voxelcount_percentages.R.
Image Analysis Pipeline: EEG Aperiodic Activity
EEG Preprocessing
EEG data were preprocessed in McKeon et al., 2024, DCN. Code and instructions for preprocessing are available (here.
EEG Source Localization and Aperiodic Activity
EEG activity was source reconstructed using each individual’s cortical, scalp, and head anatomy, resulting in processed EEG timeseries in individual HCP-MMP regions. Source localization was performed in Brainstorm with (/image_processing/EEG_aperiodic/brainstormSourceLocal.m. This script includes steps to import anatomical MRIs and preprocessed EEG, project EEG channels to the scalp, compute a three-layer forward head model, calcualte the inverse process, and average vertex-level reconstructed EEG timeseries in HCP-MMP frontal regions.
The above matlab script also contains code to compute the PSD and run FOOOF to derive the aperiodic component of the PSD and associated measures (i.e., the aperiodic exponent). Frontal lobe measures of the aperiodic exponent were examined and exclusions were performed in (image_processing/EEG_aperiodic/aperiodic_glasseratlas_cleaning.R](https://github.com/LabNeuroCogDevel/corticalmyelin_maturation/blob/main/image_processing/EEG_aperiodic/aperiodic_glasseratlas_cleaning.R).
Sample Construction
Construction of Final Sample
The initial developmental dataset considered for inclusion in this project was comprised of 264 longitudinal sessions (with correctly acquired, i.e. CuBIDS non-variant, UNI and quantitative T1 images) from 159 healthy participants. Sessions were iteratively excluded for:
- failing visual quality control of UNI and R1 images (44 sessions/18 subjects excluded)
- being collected within 6 months of another session that passed QC (4 sessions/0 subjects excluded)
- having a whole-brain mean R1 > 4 SD above the group mean (1 session/1 subject excluded)
This resulted in a final sample of 215 longitudinal sessions collected from 140 individuals, finalized in /sample_construction/finalsample_7Tmyelin.Rmd.
Generation of Final Input Dataframes
After determining the final study sample (final subject and session lists), /surface_metrics/surface_measures/extract_depthdependent_R1.R was run in order to extract depth-specific R1 measures from parquets for the final study sample and to average R1 across depths into superficial, middle, and deep cortical compartments. The extract_depthdependent_R1.R script parses parquets (generated by collate_study_anatomicalstats.py) using the flexible /surface_metrics/surface_measures/extract_surfacestats.R function and produces RDS files with R1 measures for the final sample that are used for statistical modeling. Note, averaging of R1 across depths into superficial, middle, and deep cortical compartments was informed by the results obtained below in Cortical Myeloarchitecture Validation, Characterization, and Compartments.
Statistical Analysis: Model Fitting with GAMMs
This project used generalized additive mixed models (GAMs with random intercepts) to characterize developmental change in R1 and to model associations between R1 and both EEG aperiodic activity and cognitive measures. A series of GAMM fitting functions from /gam_models/gam_functions.R were used throughout analyses, all which use gam from the mgcv package. gam_functions.R contains the following functions:
- gam.statistics.smooths: A function to fit a GAM(M) with one smooth (s(x)) plus optional id-based random effect terms and save out 1) model statistics, 2) spline-derived characteristics, 3) fitted values for the smooth from a prediction df, 4) zero-centered smooth fits, and 5) the instantaneous first derivative of the smooth. This function was used to fit most developmental models in main and sensitivity analyses
- gam.agebysex.interaction: A function to fit a GAM(M) with a factor-smooth interaction (s(x) + s(x, by = y) + y) and save out interaction statistics. This was used to fit age-by-sex interaction models
- gam.linearcovariate.maineffect: A function to fit a GAM(M) with a main smooth plus a linear covariate of interest (s(x) + y) and save out statistics for the covariate term. This was used to test for main effects of sex
- gam.covariatesmooth.maineffect: A function to fit a GAM(M) with a main smooth plus a smooth for a covariate of interest (s(x) + s(y)) and save out statistics for the covariate smooth. This was used for quantifying relationships of R1 with aperiodic activity and cognitive measures
- gam.factorsmooth.interaction: A function to fit a GAM(M) with a main smooth plus a factor-smooth interaction for a second smooth term (s(x) + s(y) + s(y, by = z)) and save out main effects and interaction results. This was used to test for superficial/deep compartment interactions
These functions were applied in a series of scripts included in the /gam_models directory to model developmental effects (/gam_models/fit_ageGAMs_glasserregions_bydepth.R and /gam_models/fit_agedepth_interactionGAMs_glasserregions.R), to conduct sensitivity analyses (/gam_models/fit_sensitivityGAMs_glasserregions_bydepth.R), to examine relationships with EEG (/gam_models/fit_EEGGAMs_glasserregions_bydepth.R, and to characterize cognitive associations (gam_models/fit_cognitionGAMs_glasserregions_bydepth).
Results: Quantitative Imaging of Myelin in Superficial, Middle, and Deep Cortical Ribbon
Cortical Myeloarchitecture Validation, Characterization, and Compartments
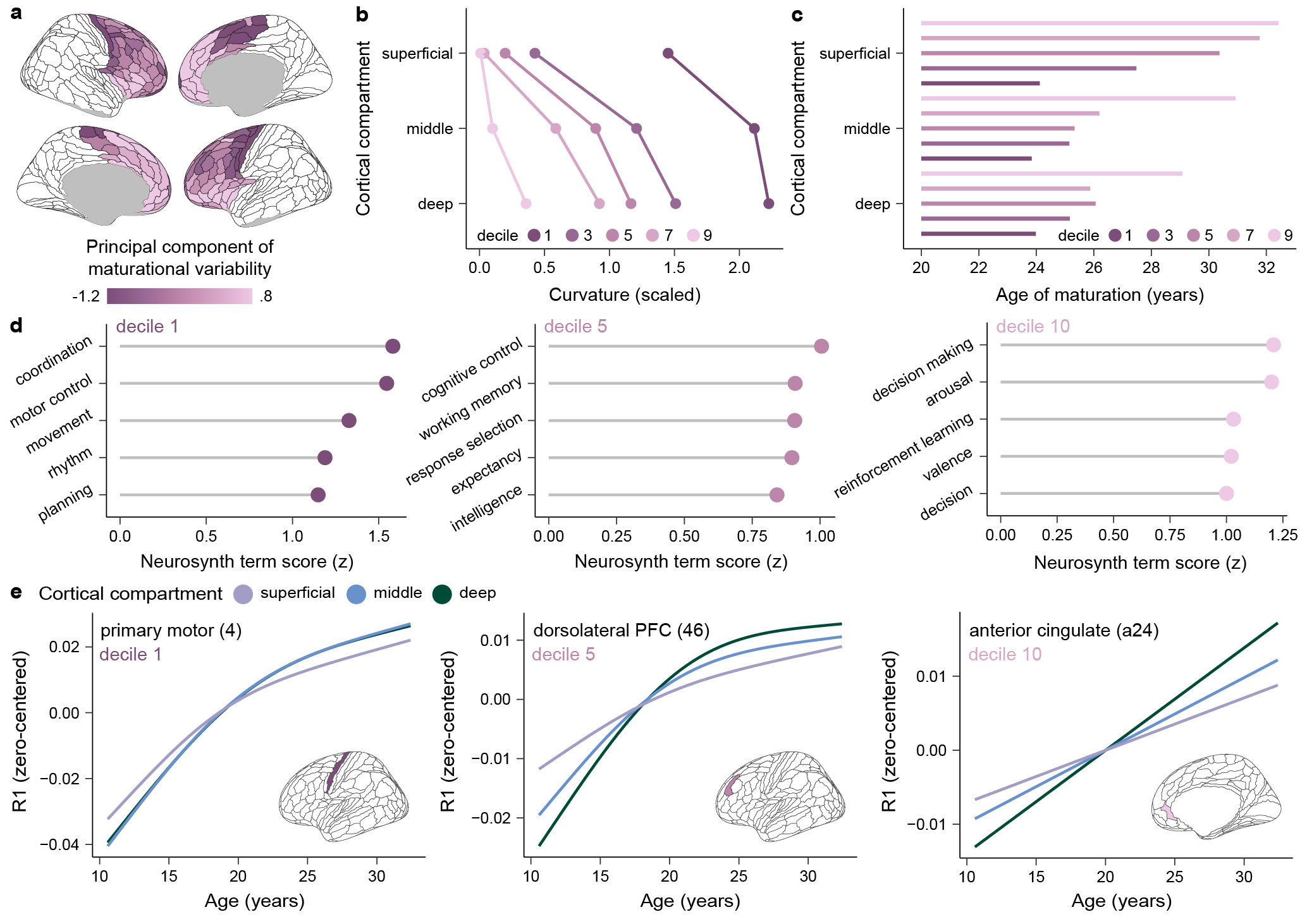
R1 was validated in our sample as a quantitative 7T imaging measure capable of capturing variation in myelin levels across cortical regions and superficial, middle, and deep cortical compartments. Characterization of R1-based myeloarchitecture was conducted in /results/R1_corticalmyelin_anatomy/R1_cortical_myeloarchitecture.Rmd. This script shows spatial variation in R1 and its alignment with a multi-modal myelin map derived from a PCA applied to a developmental T1w/T2w ratio map, a developmental MT saturartion map, and an adult myelin basic protein gene expression map. It additionally compares laminar R1 profiles and the skewness of these profiles between 1 mm developmental R1 data and data collected with 0.5 mm resolution. Finally, it demonstrates the capacity for the developmental R1 data to distinguish signal in 3 cortical ribbon compartments (superficial, middle, and deep) and uses a derivative analysis applied to the developmental and high-resolution (0.5 mm) data to identify compartment boundaries. A rendered version of this R markdown is available here!
The above code utilizes a myelin basic protein gene expression map. Myelin basic protein gene expression was obtained from vertex-level gene expression data generated from the AHBA by Wagstyl et al., 2023, elife. The script /results/R1_corticalmyelin_anatomy/myelin_maps/create_AHBAmagicc_genemap.sh gets MBP gene maps by interfacing with /results/R1_corticalmyelin_anatomy/myelin_maps/create_AHBAmagicc_genemap.py to pull gene expression and gene info csvs and produce a parcellated MBP cifti.
BrainSmash Significance
The above results quantify correlations between brain maps, including whole-brain correlations between R1 and a multi-modal myelin map and whole-brain correlations between R1 laminar profile skewness derived from 1 mm and 0.5 mm resolution data. To determine the statistical significance of these spatial correlations, autocorrelation preserving null models were constructed using BrainSmash. The procedure to compute correlation p-values with BrainSmash nulls requires 1. a parcellated distance matrix for your cortical atlas of interest, and 2. parcellated brain maps (empirical) to use for creating spatial autocorrelation-preserving surrogate maps (nulls). The distance matrix (1) was created by first deriving a vertex-wise geodesic distance matrix for left and right cortical surfaces with /results/brainsmash_nulls/compute_dense_geodesicdistmat.py and then parcellating it with the HCP-MMP atlas in /results/brainsmash_nulls/compute_parcellated_geodesicdistmat.py; a frontal lobe-only distance matrix was generated in /results/brainsmash_nulls/mask_parcellated_distmat_frontallobe.R. Surrogate maps (2) were created for whole-brain R1 and whole-brain laminar skewness using the create surrogate map python scripts.
The significance of the correlation between regional R1 and myelin PC1 was assessed using autocorrelation-preserving R1 surrogates. Visualization of example surrogates and significance testing was executed in /results/brainsmash_nulls/R1_cortical_myeloarchitecture_brainsmash.Rmd which can be viewed here.
The significance of correlations between region-wise laminar skewness in R1 in 1mm and 0.5mm data was assessed using autocorrelation-preserving surrogates as well. A rendered .html file with significance results can be viewed here.
Results: Heterochronous Laminar Myelination in the Human Prefrontal Cortical Ribbon
Heterochronous and Hierarchical Laminar Myelination
Heterochronous myelin maturation in the human frontal cortical ribbon was investigated in /results/R1_compartment_development/R1_compartmentdevelopment.Rmd. This markdown file examines trajectories of myelin development across superficial, middle, and deep cortical comparments, elucidates drivers of regional differences in the rate and timing of myelin development within cortical compartments, and functionally characterizes the principal component of maturational variability in laminar myelination trajectories. A rendered version of this R markdown is available here!
R1_compartmentdevelopment.Rmd was paired with /results/R1_compartment_development/R1_sexdifferences.Rmd, which is knitted here, to examine whether developmental trajectories differed by sex.
Results: Sensitivity Analyses
A series of sensitivity analyses were undertaken to ensure that developmental results were not driven by individual differences in sex, structural data quality, or cortical architecture. Sensitivity GAMMs were all fit in /gam_models/results/fit_sensitivityGAMs_glasserregions_bydepth.R and then examined separately in /results/R1_sensitivity_analyses scripts as explained below.
Biological Sex
Developmental models were covaried for sex and developmental results were examined in /results/R1_sensitivity_analyses/R1_sensitivity_sex.Rmd which is knitted for visualization here.
Structural Data Quality
Developmental models were covaried for euler number and developmental results were examined in /results/R1_sensitivity_analyses/R1_sensitivity_euler.Rmd which is knitted here. The euler number was computed by FreeSurfer’s mris_anatomical_stats, which is run as part of the UNI and R1 processing pipeline.
Cortical Thickness
Developmental models were covaried for regional cortical thickness and developmental results were examined in /results/R1_sensitivity_analyses/R1_sensitivity_corticalthickness.Rmd which is knitted here. Cortical thickness was computed by FreeSurfer’s mris_anatomical_stats in every HCP-MMP region for every participant.
Cortical Curvature
Developmental models were covaried for regional curvature and developmental results were examined in /results/R1_sensitivity_analyses/R1_sensitivity_curvature.Rmd which is knitted here. Curvature represents FreeSurfer’s mean curvature measure, which varies according to folding properties and gyral versus sulcal location.
Cortex Fraction (PVE)
Developmental models were covaried for regional cortex fraction (quantified at each depth) and developmental results were examined in /results/R1_sensitivity_analyses/R1_sensitivity_cortexfraction.Rmd which is knitted here. See “Determining the Cortex Volume Fraction at Each Intracortical Depth” above for scripts used to calculate depth-specific volume fractions.
Results: Cortical Myelin Supports E/I-linked Neural Dynamics
Associations Between Superficial and Deep R1 and EEG Aperiodic Activity
Relationships between superficial and deep cortex R1 and the EEG-derived aperiodic exponent were studied in /results/R1_EEG_relationships/R1_aperiodicactivity_associations.Rmd. This markdown presents the results of models examining the association between the aperiodic exponent and regional R1 and how the strength of this association differs between deep and superficial cortex. A rendered version of this R markdown is available here!
Results: Cortical Myelin Impacts Learning Rate and Cognitive Processing Speed
Associations Between Cortical R1 and Cognitive Abilities
We studied relationships between superficial and deep cortex R1 and cognitive subprocesses, including learning rates and processing speeds. Cognitive testing utilized a two-stage sequential decision-making task, an anti-saccade task, and a visually-guided saccade test (the latter 2 tests used for specificity analyses). Learning rates were obtained by fitting a 7-parameter reinforcement learning model to trial-level data on the sequential decision-making task; a seperate learning rate was obtained for each of the 2 stages of the task. Cognitive processing speed was proxied by the average across-trial response time on each of the 2 stages of the task.
Analyses examining the influence of cortical R1 on cognitive measures are presented in /results/R1_cognition_relationships/R1_cognition_associations.Rmd. This markdown explores developmental changes in learning rate and processing speed on both stages of the task and characterizes relationships between frontal lobe R1 and cognitive measures. It furthermore includes Neurosynth decoding of brain regions where myelin is linked to learning rates and a specificity analysis using anti-saccade and visually-guided saccade data. A rendered version of the html is viewable here.
SOFTWARE
Software dependencies, versions, and system requirements
- SPM BIDS app container (https://hub.docker.com/r/bids/spm/)
- FreeSurfer BIDS app container; tag freesurfer:7.4.1-202309 (https://hub.docker.com/r/bids/freesurfer)
- Neuromaps container; tag 0.0.4 (https://hub.docker.com/r/netneurolab/neuromaps)
- AFNI version 23.1.10 for Linux
- Connectome Workbench 1.5.0 (https://github.com/Washington-University/workbench/releases)
- EEG lab 2022.1 (https://eeglab.org/others/Compiled_EEGLAB.html)
- FOOOF 1.0.0 (https://fooof-tools.github.io/fooof/)
- Brainstorm 03 (https://github.com/brainstorm-tools/brainstorm3)
- BrainSMASH 0.11.0 (https://brainsmash.readthedocs.io/en/latest/index.html)
- R version 4.2.23
- hBayesDM in R (https://github.com/LabNeuroCogDevel/daw_tissue_iron/blob/main/hBayesDM_script.R)
- No non-standard hardware required